Table of Contents
Whole slide images and virtual slides
PMA.core supports a variety of image capturing techniques and modalities.
A full list of currently supported formats is available from our website at https://www.pathomation.com/formats.
You can read more about our philosophy regarding the support of vendor-specific file format in our press release.
An introduction discussing the file format issue in digital pathology is available through the Digital Pathology Today podcast.
Brightfield
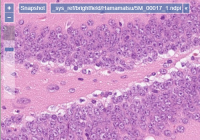
PMA.core slide reading engine supports over 35 different vendor-specific file formats that are used to encapsulate brightfield microscopy data. We offer a mix of open source community formats (OMERO TIFF & ZARR, [DICOM, …), as well as legacy ones like Olympus Webview.
A full up-to-date list is available on our website at https://www.pathomation.com/formats.
Fluorescence
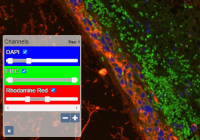
DAPI, Cy3, Cy5, … If these terms sound familiar, changed are you're engaged in fluorescent microscopy. We've been working with many vendors throughout the years to optimally support this imaging type. You can toggle individual channels directly in PMA.core, and do histogram clipping as you see fit. Advanced downstream application (that run on top of PMA.core) like PMA.studio can even visualize the histogram of each channel for you, and apply additional per-channel gamma correction.
An up-to-date list of supported fluorescent file formats is available on our website at https://www.pathomation.com/formats.
Z-stacking
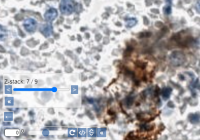
Both brightfield and fluorescent slides are often generated on a microtome, which results in smooth flat surface areas. However, when working with cytological samples, or doing microscopy in crop research, this is now always the case. Smears and similar materials are typically captured through z-stacking, whereby a sample is traversed in miniscule steps of one or two micron at the time.
An up-to-date list of z-stacking formats supported by PMA.core is available on our website at https://www.pathomation.com/formats.
Time series
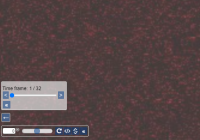
Say that you want to observe a cell culture or embryo development during a period of 24 hours. Some manufacturers allow capturing all the data from that time period in a single container (possible in combination with other dimensions like the above-mentioned).
Recently we've added support for these time-series data. We don't have too much reference datasets yet though, so if you have use cases for these, do let us know. We can then work with you to find the most optimum way of presenting these to further your specific (research) project.
Annotation formats
Because a picture says more than a thousand words, oftentimes slides get annotated. Here are some examples:
- An instructor wants to get a student's attention to a particular area on a slide by drawing arrows in various colors, possibly labeling them
- A CRO has its pathologists annotate slides for specific types of tissue like necrotic or tumor tissue. The pathologists do this according to a pre-determined color scheme
- A pharma company employs various types of image analysis tools (AI/ML/DL; all of these tools produce annotation layers that need to be integrated for further study and comparison
Pathomation's PMA.core distinguishes between 3 different kinds of annotations:
| Annotation type | Format examples | Can read | Can write |
|---|---|---|---|
| Native | MRXS, NDPI(A), SVS | Yes | No |
| PMA.core | WKT | Yes | Yes |
| Third-party | MLD, XML | Yes | No |
More information on the different kinds of annotations and how they are handled in the Pathomation platform can be found in our blog article.
Forms and meta-data
Synopic reporting a great way to structure information that was previously largely unstructured.
Here's a example: instead of letting a pathologist write a report in which he says that the patient's tissue shows signs of abnormally proliferating cell groups, you let him or her fill out a form with a checkbox for “TumorTissuePresent”.
PMA.core supports this kind of data capture and the ability to generate forms. For more information, see the specific section on the subject in this wiki.
Of course PMA.core isn't the only tool in the world that can do this. We acknowledge that by providing various options to integrate external meta-data into PMA.core as well. For more information on that subject, we refer to our blog article.
Miscellaneous
Microsoft Excel and CSV
 In PMA.core, much information is presented in the form of a list: users, slides, forms…
In PMA.core, much information is presented in the form of a list: users, slides, forms…
No matter how extended we make these list interfaces however, there's going to be a time where you think “well, if I only could re-format it like _this_, of if I could just send it off to _that_ lady…
Most views in PMA.core therefore have an Excel export button in the top-right corner that allow you to export the current data as list. Go ahead and try it, and if you don't find what you're looking for, let us know so we can incorporate it in the next version (no promises; we'll still have to evaluate the extend to which your request would be a useful feature to have for our entire user base).
Supported Codecs for WSI
The word Codec is a combination of two words “coder” and “decoder”. A piece of each word is used to create the commonly used word “Codec”. Think of this as both a coder and decoder.
Coding and decoding images is required to convert pixels into bits of information and ROIs that can be transmitted, and translated.
PMA.core currently supports the following codecs and data encoding standards:
- Blosc
- BMP
- Deflate
- iSyntax
- JPEG
- JPEG2000
- JPEGXR
- lossless
- LZW
- PNG
- RLE
More and up-to-date information on how this applies to specific file formats can be found at our website.
Troubleshooting
See our separate section.